a distill-style blog post
an example of a distill-style blog post and main elements
Scaling Vision Transformers to Gigapixel Images via Hierarchical Self-Supervised Learning
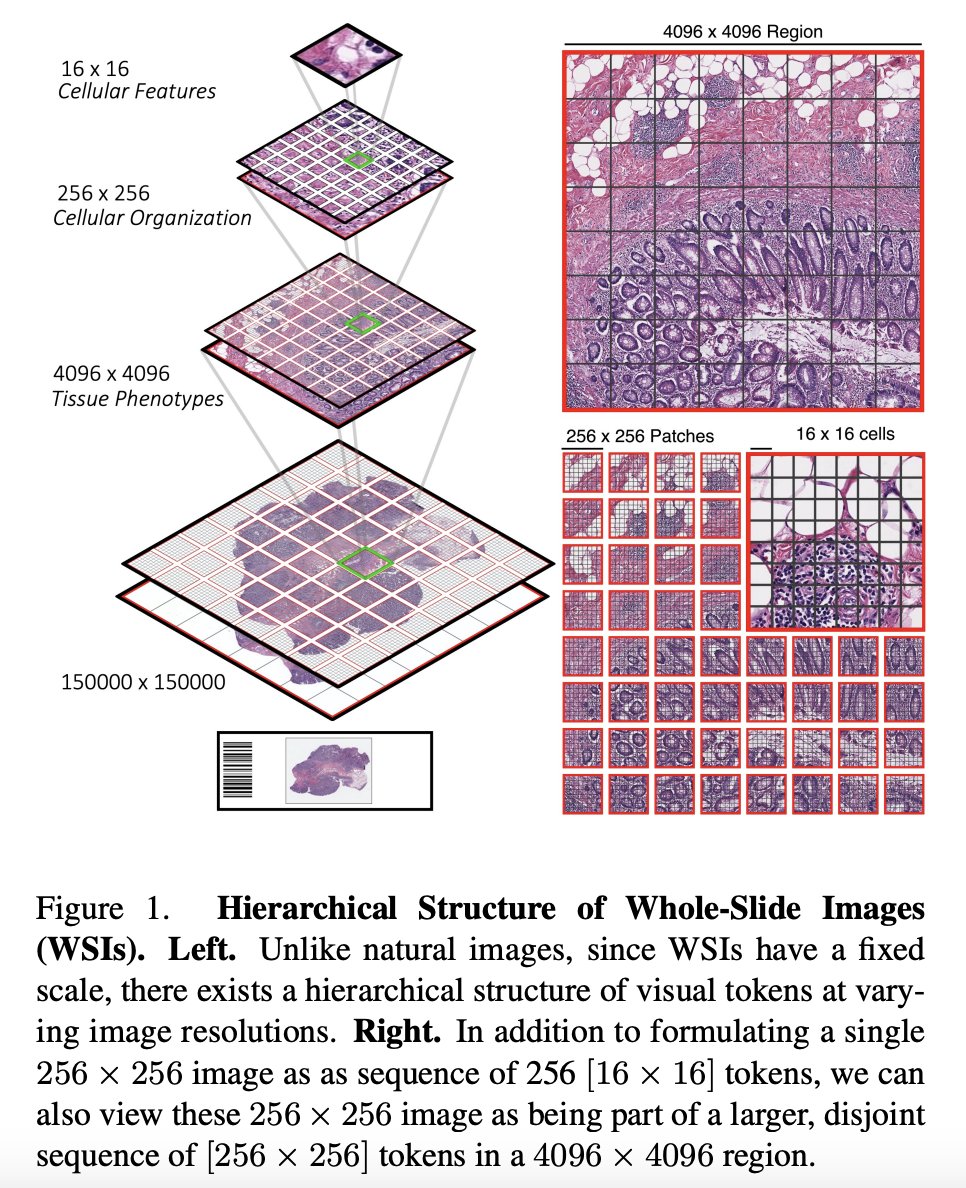
This paper proposes a new architecture for Vision Transformers (ViTs) called the Hierarchical Image Pyramid Transformer (HIPT), which is designed to handle gigapixel whole-slide imaging (WSI) in computational pathology. The authors argue that traditional ViTs are not well-suited for this task due to the large size of gigapixel images, which can contain millions of pixels. To address this challenge, HIPT uses a hierarchical structure to break down the image into smaller regions and learn representations at different levels of abstraction.
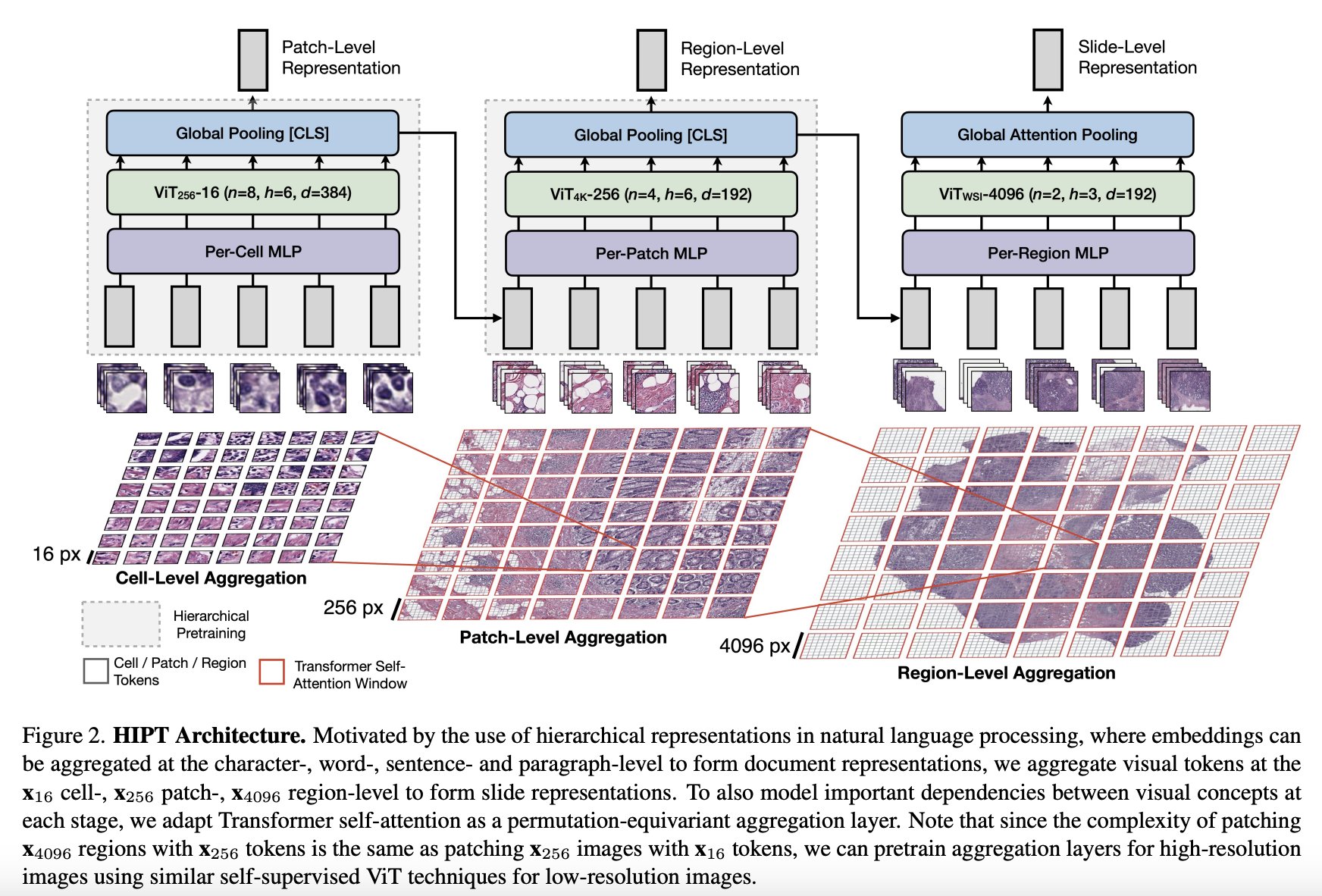
Model Overview
The model consists of three stages of hierarchical aggregation, starting with bottom-up aggregation from 16x16 visual tokens in their respective 256x256 and 4096x4096 windows to eventually form the slide-level representation.
Key Components of the HIPT Model:
-
Hierarchical Aggregation: HIPT aggregates visual tokens at the cell-, patch-, and region-level to form slide representations. This approach allows the model to capture information at different levels of granularity, from individual cells to broader tissue structures.
-
Transformer Self-Attention: To model important dependencies between visual concepts at each stage of aggregation, HIPT adapts Transformer self-attention as a permutation-equivariant aggregation layer. This enables the model to capture complex relationships and learn representations that encode both local and global context within the images.
-
Pretraining and Self-Supervised Learning: HIPT is pretrained using self-supervised learning on a large dataset of gigapixel WSIs across 33 cancer types. It leverages two levels of self-supervised learning to learn high-resolution image representations and uses student-teacher knowledge distillation for each aggregation layer.
-
Performance and Applications: HIPT with hierarchical pretraining outperforms current state-of-the-art methods on slide-level tasks. It demonstrates superior performance in capturing broader prognostic features in the tissue microenvironment, evaluated on 9 slide-level tasks including cancer subtyping and survival prediction.